Towards Out-of-core ND-Arrays -- Spilling to Disk
This work is supported by Continuum Analytics and the XDATA Program as part of the Blaze Project
tl;dr We implement a dictionary that spills to disk when we run out of memory. We connect this to our scheduler.
Introduction
This is the fifth in a sequence of posts constructing an out-of-core nd-array using NumPy, Blaze, and dask. You can view these posts here:
We now present chest a dict type that spills to disk when we run out of
memory. We show how it prevents large computations from flooding memory.
Intermediate Data
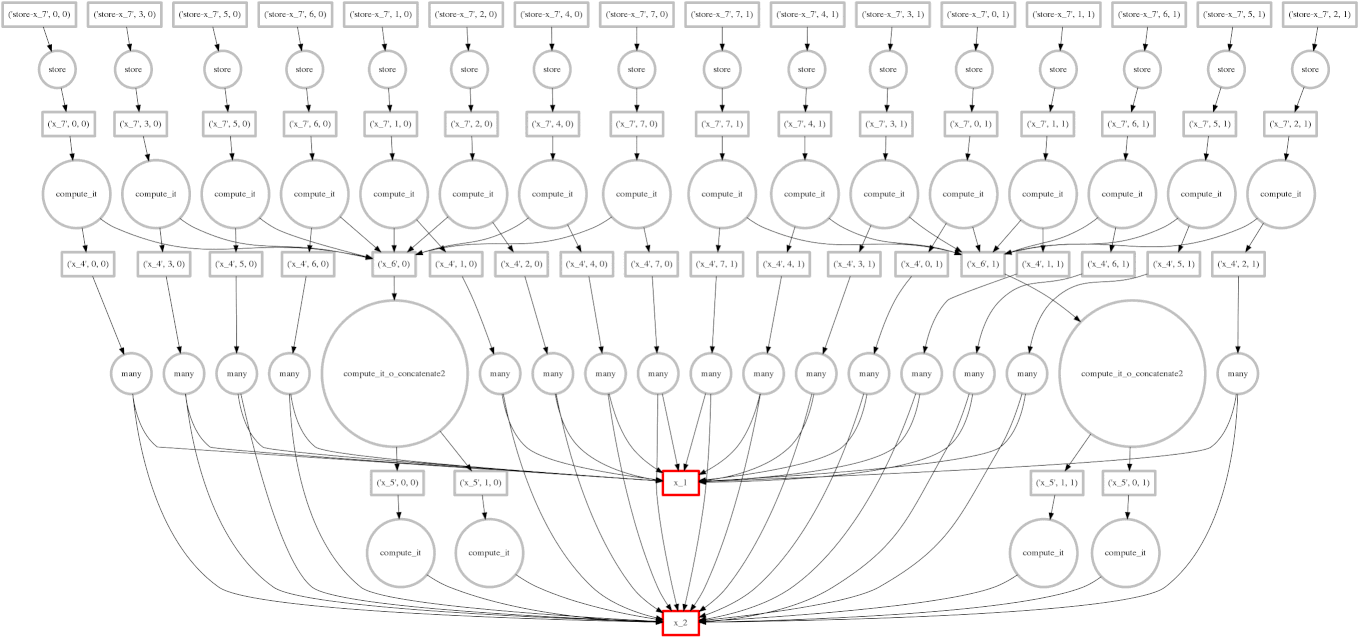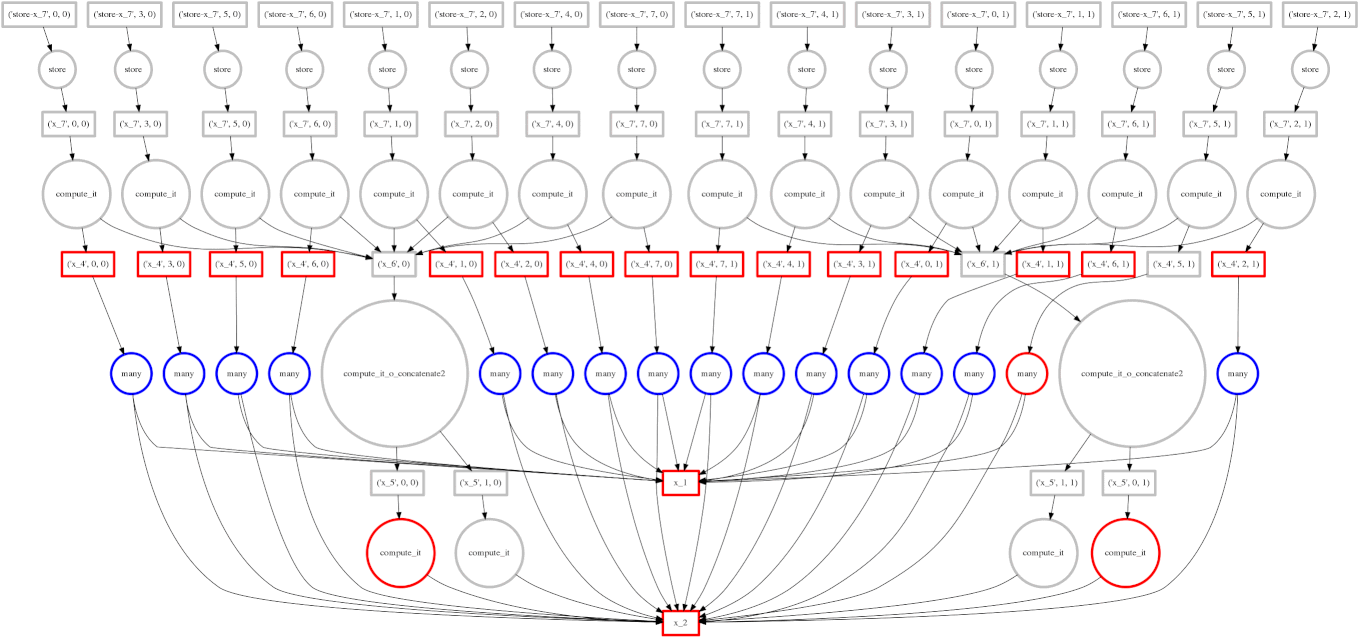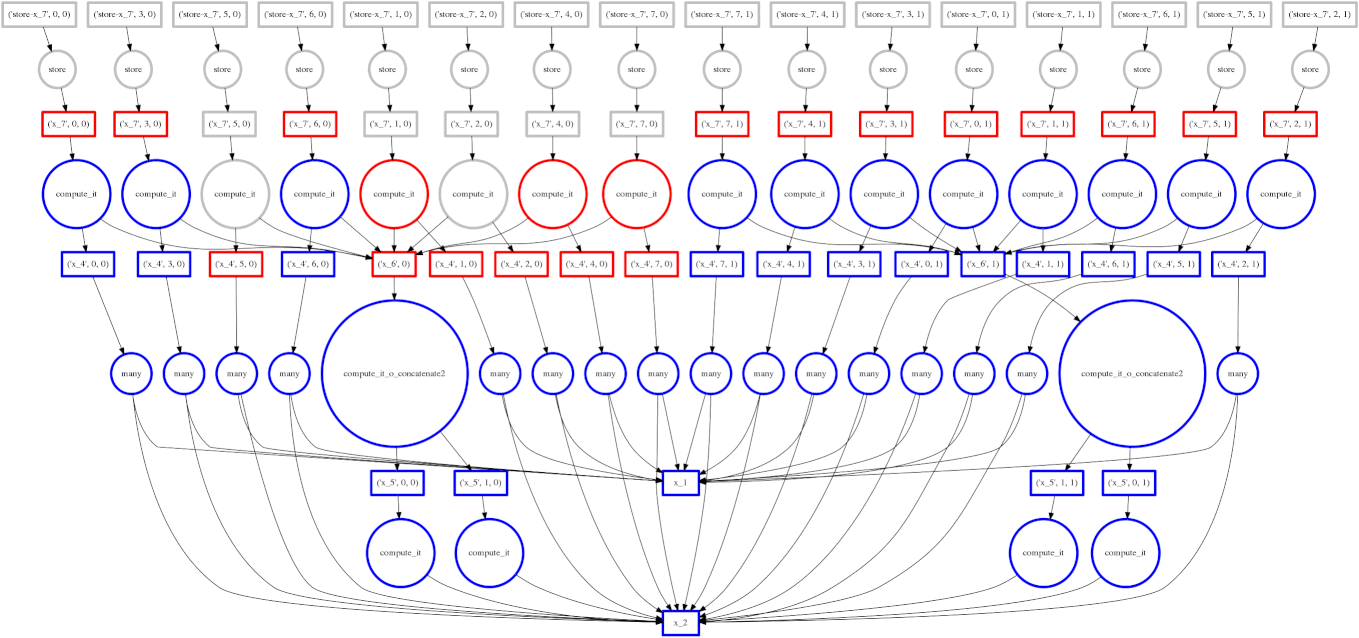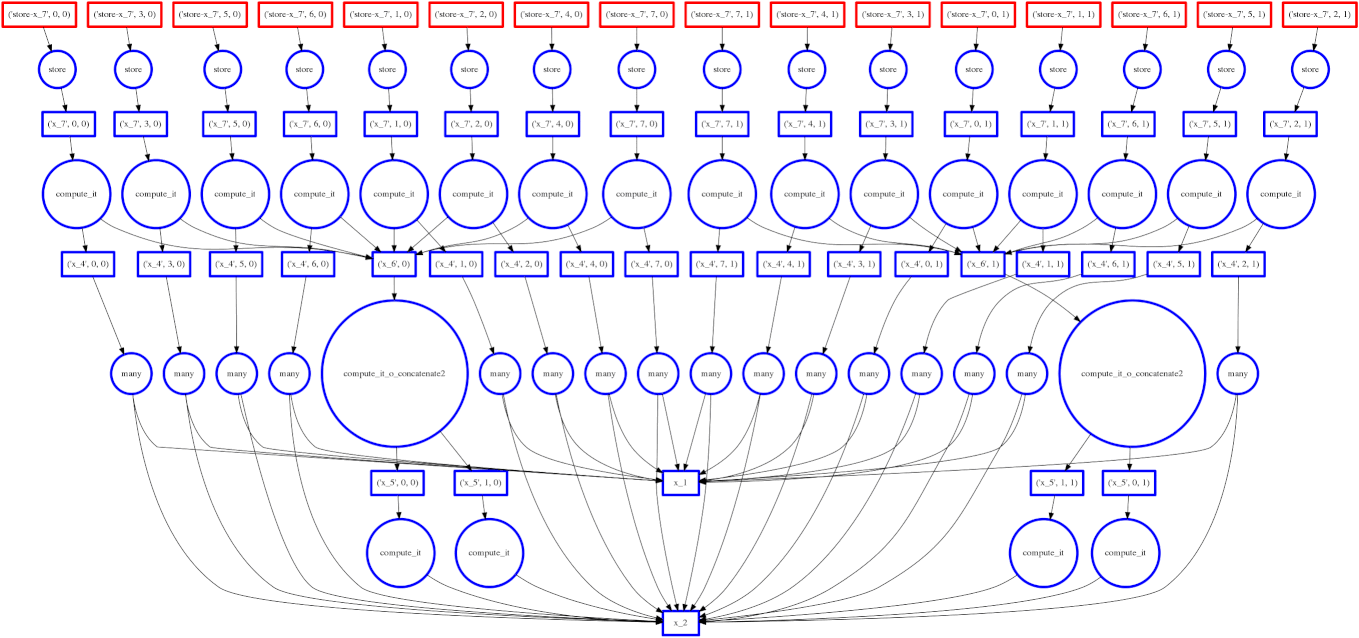
If you read the post on scheduling you may recall our goal to minimize intermediate storage during a multi-worker computation. The image on the right shows a trace of our scheduler as it traverses a task dependency graph. We want to compute the entire graph quickly while keeping only a small amount of data in memory at once.
Sometimes we fail and our scheduler stores many large intermediate results. In these cases we want to spill excess intermediate data to disk rather than flooding local memory.
Chest
Chest is a dict-like object that writes data to disk once it runs out of memory.
>>> from chest import Chest
>>> d = Chest(available_memory=1e9) # Only use a GigaByte
It satisfies the MutableMapping interface so it looks and feels exactly like
a dict. Below we show an example using a chest with only enough data to
store one Python integer in memory.
>>> d = Chest(available_memory=24) # enough space for one Python integer
>>> d['one'] = 1
>>> d['two'] = 2
>>> d['three'] = 3
>>> d['three']
3
>>> d.keys()
['one', 'two', 'three']
We keep some data in memory
>>> d.inmem
{'three': 3}
While the rest lives on disk
>>> d.path
'/tmp/tmpb5ouAb.chest'
>>> os.listdir(d.path)
['one', 'two']
By default we store data with pickle but chest supports any protocol
with the dump/load interface (pickle, json, cbor, joblib, ….)
A quick point about pickle. Frequent readers of my blog may know of my
sadness at how this library
serializes functions
and the crippling effect on multiprocessing.
That sadness does not extend to normal data. Pickle is fine for data if you
use the protocol= keyword to pickle.dump correctly . Pickle isn’t a good
cross-language solution, but that doesn’t matter in our application of
temporarily storing numpy arrays on disk.
Recent tweaks
In using chest alongside dask with any reasonable success I had to make the
following improvements to the original implementation:
- A basic LRU mechanism to write only infrequently used data
- A policy to avoid writing the same data to disk twice if it hasn’t changed
- Thread safety
Now we can execute more dask workflows without risk of flooding memory
A = ...
B = ...
expr = A.T.dot(B) - B.mean(axis=0)
cache = Chest(available_memory=1e9)
into('myfile.hdf5::/result', expr, cache=cache)
Now we incur only moderate slowdown when we schedule poorly and run into large quantities of intermediate data.
Conclusion
Chest is only useful when we fail to schedule well. We can still improve
scheduling algorithms to avoid keeping data in memory but it’s nice to have
chest as a backup for when these algorithms fail. Resilience is reassuring.
blog comments powered by Disqus